The Challenge¶
Alzheimer's disease and Alzheimer's disease-related dementias (AD/ADRD) are a group of brain disorders characterized by progressive cognitive impairments that severely impact daily functioning. Early prediction of AD/ADRD is crucial for potential disease modification through emerging treatments, but current methods are not sensitive enough to reliably detect the disease in its early or presymptomatic stages.
The PREPARE: Pioneering Research for Early Prediction of Alzheimer's and Related Dementias EUREKA Challenge sought to advance accurate, innovative, and equitable solutions for early prediction of AD/ADRD. To achieve this goal, the challenge featured three phases that built successively on one another. This blog post shares the results and winners of the third and final phase.
| Phase | Description |
|---|---|
| Phase 1 Find IT! |
Solvers from across academia and industry found, curated, or contributed representative and open datasets that can be used for early prediction of AD/ADRD across a range of modalities including neuroimagery, synthetic electronic health records, survey, speech, and mobile apps. Read more about the results of Phase 1 here. |
| Phase 2 Build IT! |
Data science solvers advanced algorithms and analytic approaches for early prediction of AD/ADRD, with an emphasis on explainability of predictions, in two data tracks focused on acoustic voice data and social determinants of health survey data. Read more about the results of Phase 2 here. |
| Phase 3 Put IT All Together! |
Ten top teams from Phase 2 refined their algorithmic approaches, working to make their solutions more rigorous and generalizable in a real-world context. Included a public virtual pitch event, and an in-person winner showcase at the NIH. |
Phase 3 brought together the potential of winning data and models from the previous phases, advancing toward a future where machine learning and rich research data can identify patients at risk of developing AD/ADRD earlier and more accurately.
An expert panel of NIH judges selected the winning teams based on their outstanding innovation in early AD/ADRD prediction, scientific rigor, models that generalize well across diverse populations, and clear communication of insights to benefit the broader research community. The winning solutions represent the culmination of months of work on data integration, model refinement, and rigorous experimentation, but also the contributions of hundreds of researchers and participants worldwide who created and shared the data sources that made the PREPARE Challenge possible.
Results¶
All ten qualifying teams participated in Phase 3. Teams represented a range of backgrounds and disciplines, from computer science and engineering to biomedical research and health sciences. Over the course of several months, participating teams made their prize-winning Phase 2 models more rigorous, generalizable, and useful for the field, incorporating new data from underrepresented populations, correcting for overfitting, identifying and mitigating bias, testing experimental biomarkers, and improving model explainability.
Solvers shared their findings in two dissemination events, to align with the challenge goal of sharing insights with the research community.
- In a public virtual pitch event, all participating teams were invited to give brief talks on what they did and what they found. The insights shared remain available for the broader research community via a public recording of the pitch event.
- In an in-person winner showcase event at the NIH, representatives from the winning teams met with judges, challenge organizers, and stakeholders within and outside of NIA to give talks and celebrate the competition results.
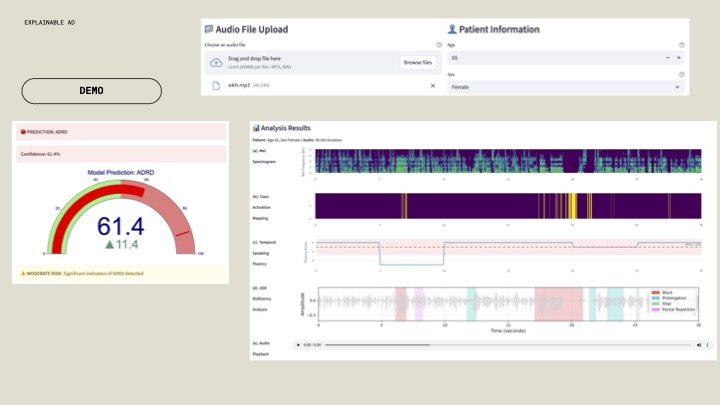
Winner Overview¶
| Prize | Winner | Approach Highlights |
|---|---|---|
| 1st Place |
RASKA-Team
(Social Determinants)
Social scientists with expertise in economics, statistics, epidemiology, and NLP
|
Harmonized data from four additional countries in the HCAP network, creating a model that could be used more broadly and enabling thorough and rigorous fairness and generalizability testing. |
| 2nd Place |
ExplainableAD
(Acoustic)
Engineers who specialize in analyzing medical image and audio data
|
Used non-native and corpus-diverse speech from corpora in DementiaBank, together with a non-native English corpus, Mispeech/Speechocean762, to train a disfluency model and build an explainable clinical demo. |
| Runner-up |
NxGTR
(Social Determinants)
A computer scientist with broad ML expertise
|
Developed a robust model architecture for different kinds of research, including HCAP and NACC data. Drew specific, actionable insights on detecting cognitive decline. |
| Runner-up |
Harris and Kielo
(Acoustic)
Team combining computer science and communication expertise
|
Incorporated new healthy-speech data and demonstrated an innovative technique to generate synthetic training data based on over 60 known linguistic symptoms of ADRD. |
Clean code bonus prize winners:
The following teams earned Clean Code Bonus Prizes for the clarity, reproducibility, and usability of their code.
- RASKA (Social Determinants of Health), the first-place prize winning team of social scientists
- SpeechCARE (Acoustic), an academic research lab focused on cognitive impairment detection.
- IGCPharma (Acoustic), a clinical-stage biotechnology company.
Approaches used¶
In Phase 3, approaches were as varied as the teams themselves, reflecting a rich methodological landscape. Below, we highlight several representative top approaches from each track.
Acoustic Track
Teams from the Acoustic Track had originally worked with a subset of data from the DementiaBank, an open database aggregating spontaneous speech recordings from multiple dementia studies, to predict a combined diagnosis label (control, MCI, or AD/ADRD). Through experimentation and refinement, Phase 3 teams identified several key methodological insights about extracting signal from voice recordings:
- Integrating linguistic features (what was spoken), phonetic features (how it was spoken), and demographic metadata within multi-task frameworks enabled models to capture a broader range of cognitive signals. Pipelines based on pre-trained models such as Whisper and wav2vec provided complementary representations that improved diagnostic performance.
- Generating synthetic speech data and incorporating non-clinical speech samples from Mozilla Common Voice and non-native English speakers addressed data scarcity and supported the development of novel fluency biomarkers. These approaches expanded the training data available for model development and improved robustness.
- Adopting modular frameworks for feature extraction, harmonization, and uncertainty estimation improved models’ ability to generalize across datasets. Incorporating additional
- DementiaBank resources, including the newly released Delaware corpus, further broadened the data sources used for training and enhanced cross-corpus performance.
Social Determinants Track
Teams from the Social Determinants Track had originally worked with longitudinal survey data from the Mexican Health and Aging Study (MHAS) and its Cognitive Aging Ancillary Study (Mex-Cog) where the label was respondents' total composite score in the second included wave (a constructed MHAS variable). In Phase 3, teams identified effective strategies for using open social determinants data for early prediction of cognitive decline:
- Incorporating data from additional countries in the HCAP network improved model generalizability by accounting for differences in cognitive decline patterns across locations and communities, underscoring the importance of representative training data.
- Engineering features that captured socioeconomic and demographic factors, in addition to health-related factors, improved predictive performance. Education consistently emerged as a strong contributor across models and populations, alongside community context, family structure, and income.
- Applying boosting models enhanced with fairness adjustments and uncertainty estimation led to more reliable predictions and highlighted differences in model performance across subgroups.
- Developing generalized frameworks that operated across diverse datasets improved robustness and clinical relevance, supporting transferability across cohorts with different underlying population characteristics.
Across tracks, the challenge prompted participants to focus on methods that supported real-world use, such as improving interpretability, addressing fairness concerns, and ensuring clinical relevance, rather than optimizing for a single evaluation metric. This emphasis shaped the modeling approaches and led to notable changes in the structure and capabilities of multiple winning solutions.
Congratulations to the winners and everyone whose work was celebrated throughout this challenge. This multi-year effort brought together scientists and developers, NIA staff and judges, data contributors, and stakeholders across the AD/ADRD community who accomplished something remarkable together. Meet the winning teams below and see for yourself how they advanced early prediction in Phase 3.
Meet the Winners¶
All winning reports can be found in the winners repository on GitHub. Submissions are licensed under the open-source MIT license. If a team chose to share their solution code publicly, we've included a link in their introduction below and in the winner's respository.
RASKA-Team¶
Social Determinants of Health



Team members: Suhas D. Parandekar, Ekaterina Melianova, and Artem Volgin
Place: 1st Overall, Clean Code Bonus
Public repository: https://github.com/artvolgin/prepare-p3
Prize: $100,000 + $7,500
Hometowns: Washington, DC and London, UK
Usernames: artvolgin, Amelia, sparandekar
Background:
Suhas D. Parandekar is a Senior Economist at the World Bank’s Global Education Practice, where he leads cross-functional teams on projects in education, science, technology, and innovation.
Ekaterina Melianova recently completed a PhD in Advanced Quantitative Methods at the University of Bristol, UK. Her work focuses on applying innovative statistical methods to real-world social challenges, particularly in social epidemiology, health, and education.
Artem Volgin recently completed a PhD in Social Statistics at the University of Manchester, UK. His research focuses on applications of Network Analysis and Natural Language Processing, and he has extensive experience working with real-world data across diverse domains.
Summary of approach:
The team applied machine learning to harmonized tabular HCAP datasets from Mexico, India, the UK, and the US, aligning survey waves to create cross-nationally comparable measures of cognitive functioning. Their pipeline included data harmonization, engineered group-level statistics, and fairness adjustments for subgroup differences. Predictions were generated using LightGBM, CatBoost, and XGBoost, optimized with hyperparameter tuning, with interpretability supported through SHAP values and uncertainty quantified using Jackknife+. The final ensemble combined outputs from all three models.
Check out RASKA-team's full submission in the challenge winners repository.
ExplainableAD¶
Acoustic

Team members: Yinglu Deng and Yang Xu
Place: 2nd Overall
Prize: $50,000
Hometowns: San Diego, California and LiShui ZheJiang, China
Usernames: sheep, cecilia12312
Social Media: /in/yinglu-cecilia-deng
Background:
We are experienced engineers at a medical company, all holding Master of Science degrees. Our professional work involves processing and analyzing medical data, particularly focusing on image and audio data. Beyond our primary roles, we are enthusiastic participants in data science competitions and have achieved multiple victories in these contests.
Summary of approach:
The team developed a multi-task model based on OpenAI’s Whisper encoder, with one head for classifying diagnosis (Healthy, MCI, ADRD) and another for temporal segmentation to detect disfluencies. Training drew on nine datasets, with evaluation across four external English-language corpora using a leave-one-out approach. They introduced three fluency biomarkers: ASR-derived disfluency counts, word token probabilities, and transferred fluency scores, each showing differences between healthy and ADRD groups. An expert-labeled dataset from non-native English speakers was used to train the disfluency module (speechocean762). The characteristics of disrupted speech provided a robust signal, even when the cause of the disruption was different.
Check out ExplainableAD's full submission in the challenge winners repository.
NxGTR¶
Social Determinants of Health

Team members: Carlos Huertas
Place: Runner-up
Prize: $25,000
Hometown: Seattle, WA
Username: NxGTR
Social Media: in/carloshuertasgtr/
Background:
I am originally from Tijuana Mexico. I studied Computer Science and really enjoyed the AI space, although hardware resources were always limited, which shaped my push to always simplify. Following this path, I completed my PhD in the area of feature selection for high-dimensional spaces. Currently I work for Amazon as an Applied Scientist where we develop Machine Learning technology to protect Amazon and customers from fraudulent activity.
Summary of approach:
Created ADRDModel, a framework designed to operate across multiple datasets without requiring dataset-specific adjustments. The framework includes a Feature Fetch module for automated feature extraction and a DatasetAdapter for harmonization of study-specific data such as MHAS and NACC-UDS. Built-in uncertainty estimation provides multiple perspectives on each patient, and outputs include both predicted future cognitive scores and a personalized Health Score. Parameter optimization was performed to balance explainability and performance while maintaining model flexibility.
Check out NxGTR's full submission in the challenge winners repository.
Harris and Kielo¶
Acoustic


Team members: Matthew Kielo and Harris Alterman
Place: Runner-up
Prize: $25,000
Public repository: https://github.com/mkielo3/prepare3
Hometowns: Kingston, Ontario and Denver, Colorado
Usernames: harrisalterman, mkielo
Background:
Matthew Kielo is currently completing a Master’s in Computer Science from Georgia Tech and has spent his professional career working in quantitative finance.
Harris Alterman has a bachelor’s degree in arts with a focus on communication from the University of Waterloo. He currently works as a content creator in New York and has an avid interest in all forms of communication.
Summary of approach:
The team refactored their model around three types of information in clinical speech data: linguistic (what is said), paralinguistic (how it is said), and demographic (who is speaking). The framework identifies linguistic and paralinguistic features while adjusting for demographic and sampling effects. External data sources included the Mozilla Open Voice dataset, used to model healthy speech patterns, and a synthetic dataset created with ChatGPT transcripts informed by ADRD research to capture additional linguistic features.
Check out Harris and Kielo's full submission in the challenge winners repository.
Meet the Clean Code Bonus Prize Winners¶
SpeechCARE¶
Acoustic

Team members: Maryam Zolnoori, Hossein Azad Maleki, Ali Zolnour, Yasaman Haghbin, Sina Rashidi, Mohamad Javad Momeni Nezhad, Fatemeh Taherinezhad, and Maryam Dadkhah
Award: Clean code bonus
Prize: $7,500
Hometown: New York, NY
Social media: in/maryam-zolnoori-9aabab231/
Usernames: maryamzolnoori, yasaman.haghbin, sinarashidi
Background:
SpeechCARE team has extensive experience developing speech processing pipelines for cognitive impairment detection. Given the variability in speech corpus characteristics (e.g., speech tasks, languages) and audio quality, no single approach consistently yields optimal performance. Therefore, our team is actively exploring and refining diverse methodologies to address the critical unmet need for early detection of cognitive impairment. Our team is based out of Columbia University Irving Medical Center.
Summary of approach:
The team developed a speech processing pipeline covering eight areas: preprocessing, modeling, explainability, trustworthiness, generalizability, multi-task learning, continual learning, and application in real-world care settings. The pipeline incorporates methodological components for speech data preparation and modeling, as well as mechanisms for adapting to new data, improving interpretability, and supporting deployment in clinical contexts.
Check out SpeechCARE's full submission in the challenge winners repository.
IGC Pharma¶
Acoustic




Acoustic
Team members: Ram Mukunda, Paola Ruíz Puentes, Nestor González, and Daniel Crovo
Place: Clean code bonus
Award: $7,500
Hometowns: Potomac, MD; Bogotá, Colombia; and Washington, D.C.
Usernames: IGCPHARMA, PaolaRuiz-IGC, ngonzalezigc, dcrovo.
Social media: Ram Mukunda, Paola Ruiz Puentes, Nestor Gonzalez, Daniel Crovo
Background:
All team members work at IGC Pharma . IGC Pharma is at the forefront of clinical-stage biotechnology, passionately dedicated to revolutionizing Alzheimer’s disease treatment. In addition to our therapeutic advancements like IGC-AD1, we leverage cutting-edge AI models to predict early biomarkers for Alzheimer’s and optimize clinical trials. Our AI research also investigates potential interactions with receptors like GLP-1, GIP, and CB1.
Ram is the passionate CEO of IGC Pharma, whose decade-long journey in medical and pharmaceutical research reflects his commitment to transforming the lives of those affected by Alzheimer's disease. Paola manages the company's AI/ML team and works on innovative ML tools for AD/ADRD diagnosis. Nestor focuses on developing and refining machine learning and deep learning models and optimizing state-of-the-art large language models. Daniel Crovo, an Electronics Engineer, brings his passion for applying artificial intelligence to medical research, focusing on early detection of Alzheimer’s disease.
Summary of approach:
The team built a multilingual speech classification pipeline using a fine-tuned Whisper Large encoder, chosen for its balance of accuracy and generalizability. Training data came from 12 DementiaBank studies, with preprocessing to isolate participant speech and enhance audio quality. They froze most encoder layers to adapt to limited data and segmented recordings into 30-second chunks, averaging predictions at the participant level. To evaluate generalization, they applied two leave-one-out protocols: one holding out entire languages (English, Spanish, Mandarin) and another holding out speech tasks (image description, word fluency, discourse). They also developed a speech translation pipeline to convert non-English recordings into English while preserving acoustic features, extending the reach of English-trained models to multilingual data.
Check out IGCPharma's full submission in the challenge winners repository.
Thanks you to all challenge participants and to our incredible winners! We also thank our data partners MHAS and DementiaBank, judges, and challenge team. We thank the National Institute on Aging (NIA), an institute of the National Institute of Health (NIH), for sponsoring this challenge!